Plot decision curve stability across bootstrap replicates
Source:R/stability_plot.R
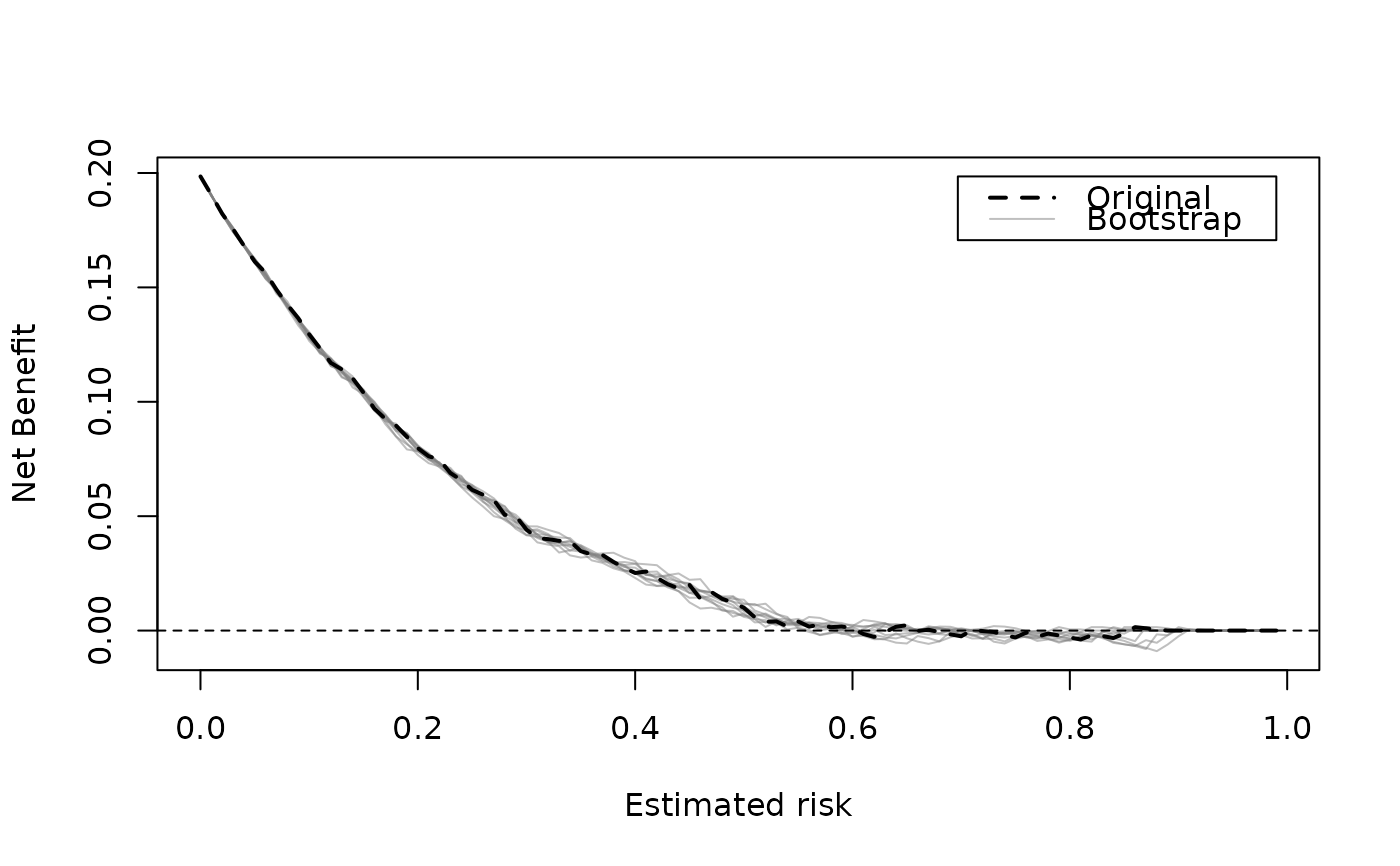
dcurve_stability.RdA decision curve (in)stability plot shows decision curves for bootstrap models evaluated on original outcome. A stable model should produce curves that differ minimally from the 'apparent' curve. See Riley and Collins (2023).
Usage
dcurve_stability(
x,
thresholds = seq(0, 0.99, by = 0.01),
xlim,
ylim,
xlab,
ylab,
col,
subset,
plot = TRUE
)Arguments
- x
an object produced by
validatewith method = "boot_*" (orboot_optimismwith method="boot")- thresholds
points at which to evaluate the decision curves (see
dcurves::dca)- xlim
x limits (default = range of thresholds)
- ylim
y limits (default = range of net benefit)
- xlab
a title for the x axis
- ylab
a title for the y axis
- col
color of points (default = grDevices::grey(.5, .5))
- subset
vector of observations to include (row indices). This can be used to select a random subset of observations.
- plot
if FALSE just returns curves (see value)
Value
plots decision curve (in)stability.
Invisibly returns a list containing data for each curve. These are returned from dcurves::dca.
The first element of this list is the apparent curve (original model on original outcome).
References
Riley, R. D., & Collins, G. S. (2023). Stability of clinical prediction models developed using statistical or machine learning methods. Biometrical Journal, 65(8), 2200302. doi:10.1002/bimj.202200302
Examples
# \donttest{
set.seed(456)
# simulate data with two predictors that interact
dat <- pmcalibration::sim_dat(N = 2000, a1 = -2, a3 = -.3)
mean(dat$y)
#> [1] 0.1985
dat$LP <- NULL # remove linear predictor
# fit a (misspecified) logistic regression model
m1 <- glm(y ~ ., data=dat, family="binomial")
# internal validation of m1 via bootstrap optimism with 10 resamples
# B = 10 for example but should be >= 200 in practice
m1_iv <- validate(m1, method="boot_optimism", B=10)
#> It is recommended that B >= 200 for bootstrap validation
dcurve_stability(m1_iv)
 # }
# }